circRNADisease v2.0 Tutorial |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Updated time: October 2023 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
[1] Overview |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Circular RNAs (termed as circRNAs), as a naturally occurring large family of non-coding RNA molecules, have been reported pervasively transcribed in eukaryotes. These single-stranded RNA molecules are characterized by covalently closed loop structures (termed as "backsplice") in which a downstream 3' splice site interacts with an upstream 5' splice site in a linear pre-messenger RNA. In recent years, increasing evidence is pointing towards circRNAs as regulators of most of cellular process, including pluripotency, epithelial-mesenchymal transition, and neural development and differentiation. Moreover, circRNAs are much more stable than linear RNAs, evolutionary conserved, and have specific spatiotemporal expression patterns, suggesting that circRNAs might involve in different biological roles. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The circRNADisease v2.0 is a database for experimentally supported circRNA dysregulations in human diseases. First, we performed an extensive literature query of the PubMed database (https://pubmed.ncbi.nlm.nih.gov/) using a list of keywords combining 'circular RNA' or 'circRNA' with 'disease', 'tumor', 'cancer', 'carcinoma', 'syndrome' or 'inflammation' mainly from January 2018 to August 2023. We downloaded all published literature involving the circRNA-disease relationships. Second, we manually extracted experimentally supported circRNA-disease associations. Different researchers were assigned to double-check all circRNA-disease pairs. Each entry in the circRNADisease v2.0 includes detailed information on a circRNA-disease association, including circRNA (circBase ID, name, and synonym), disease basic information, the circRNA expression pattern (eg. up-regulated or down-regulated), experimental detection techniques (eg. qRT-PCR, microarray, and/or sequencing), circRNA associated genes (transcription factors, genes, and/or proteins) and microRNAs, biological functions (eg. proliferation, migration, and invasion) and molecular mechanisms (eg. ceRNA, signaling pathways) of circRNA in disease, association with patients survival, a brief functional and expression description about circRNA, literature references and other annotation information. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
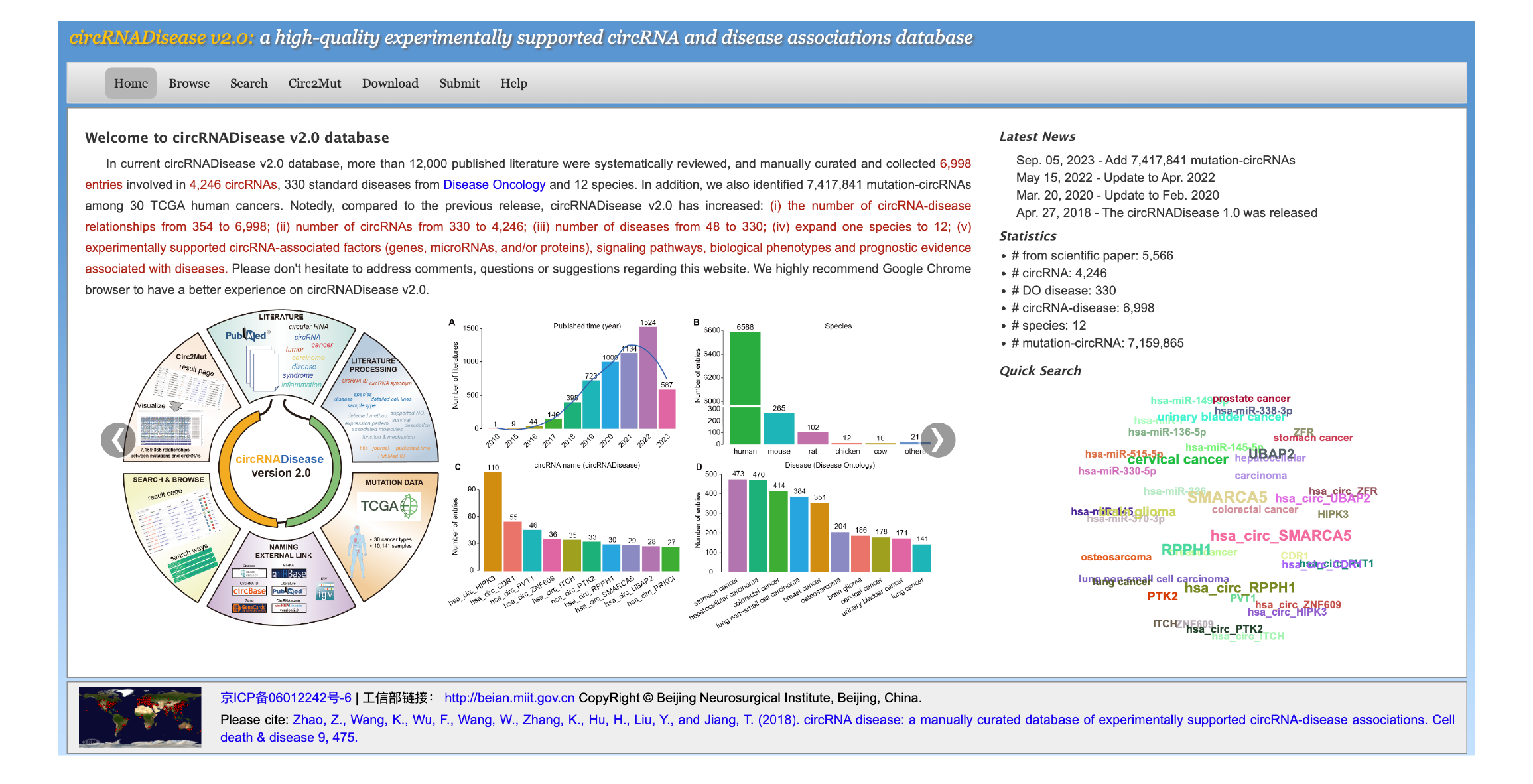
In current circRNADisease v2.0 database, 11,419 published literature were systematically reviewed, and manually curated and collected 6,998 entries involved in 4,246 circRNAs, 330 standard diseases from Disease Oncology and 12 species (human, mouse, boars, C. elegans, chicken, cow, goat, pig, rat, sheep, virus, apostichopus japonicus), supporting circRNA-disease associations from PubMed database. Since the naming of circRNA in different studies is often ambiguous and lacks consistency, we provided naming conventions and use "circ" plus gene name for uniform naming, such as circ_HIPK3, circ_ITCH, and circ_FOXO3, etc. We have organized web interface of circRNADisease 2.0 according to the three main functional features: (i) Home, (ii) Browse, (iii), Search, (iv) Download, and (v) Submit. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The circRNADisease 2.0 includes five distinctive features:
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
[2] Data statistics |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
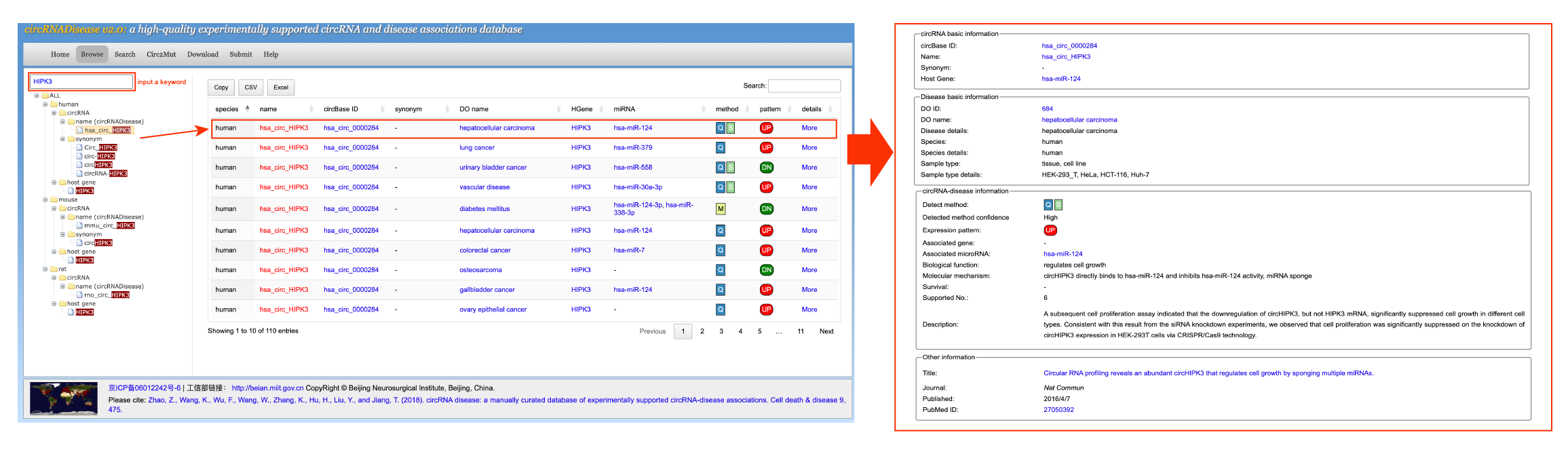
[3] BrowseTo browse circRNA-disease association data in the database, please select the menu "Browse" (Figure 1). Users can browse all the entries for different species in three ways: by circRNA, by host gene or by DO disease. Take a browse human circ_HIPK3-related entries as an example. To browse the entries for human circ_HIPK3, please click "human" -> "circRNA" -> "name (circRNADisease)" -> "circ_HIP3" option. To facilitate the user's search for human circ_HIPK3, we have added a quick fuzzy search tool in the 'Browse' module. The browse result will be displayed in the right panel. Users can browse the details by clicking the 'more' button. In the detailed section, it will be shown the circRNA basic information (including circBase ID, name, synonym, and host gene), disease basic information (including disease, species, and tissue/cell/PDC information), circRNA-disease information (detected method, expression pattern in disease, associated genes and microRNAs, biological function, molecular mechanism, survival evidence, and description), and other information (including article title, published journal and time, and PubMed ID).
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
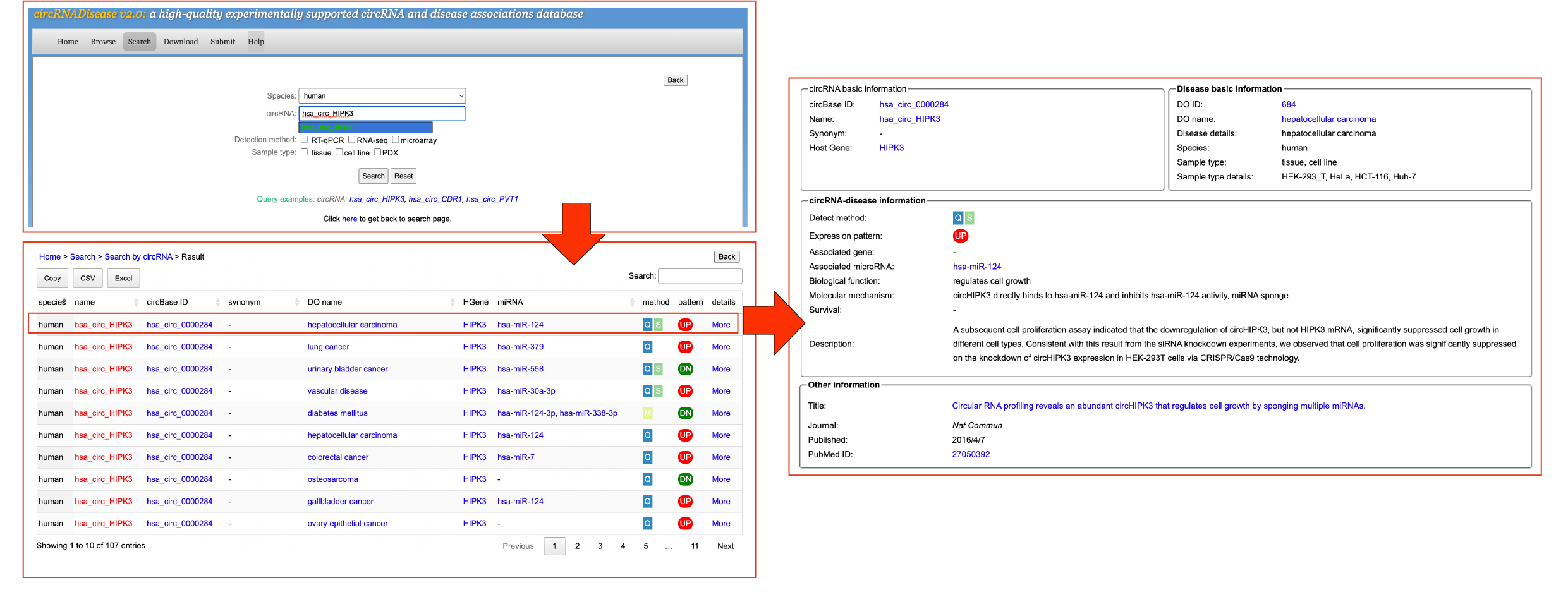
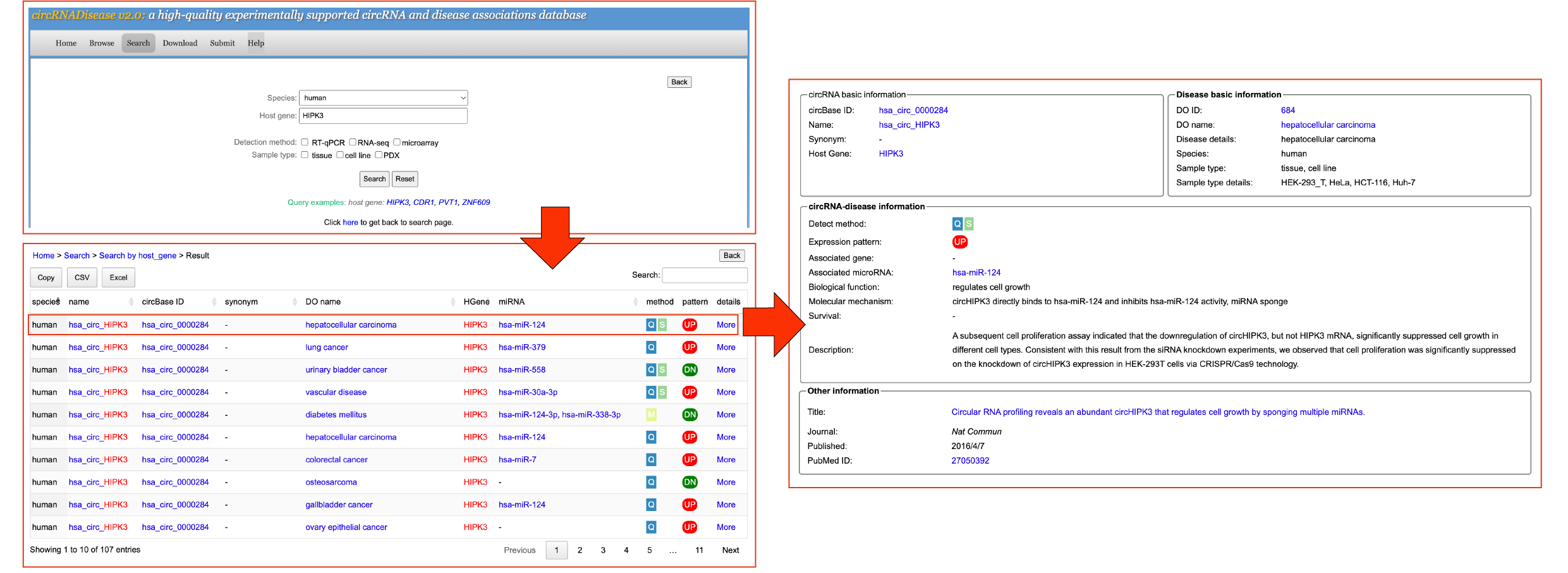
[4] SearchTo search data in the database, please select the menu "Search".
In the circRNADisease v2.0 database, users can search all entries in three ways: by circRNA, by circRNA host gene, by disease or by miRNA. Detailed information shown in search results:
[4.1] Search by circRNAUsers can click the 'Search By circRNA' button and input a circRNA name/circBase ID/synonym to search the circRNA-disease information. Take a search "human circ_HIPK3"-related entries as an example.
[4.2] Search by host geneUsers can click the 'Search by host gene' button and input a host gene to search the circRNA-disease information. Take a search "human HIPK3"-related entries as an example.
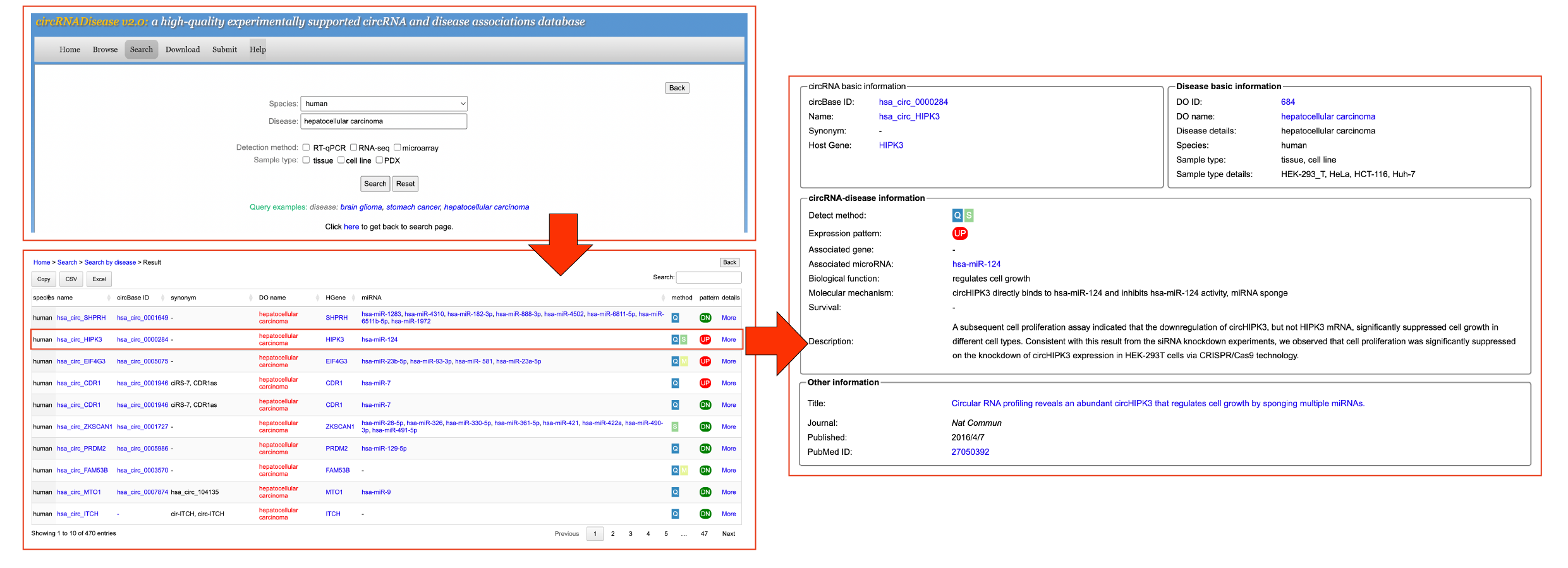
[4.3] Search by diseaseUsers can click the 'Search by disease' button and input a disease to search the circRNA-disease information. Take a search "human hepatocellular carcinoma"-related entries as an example.
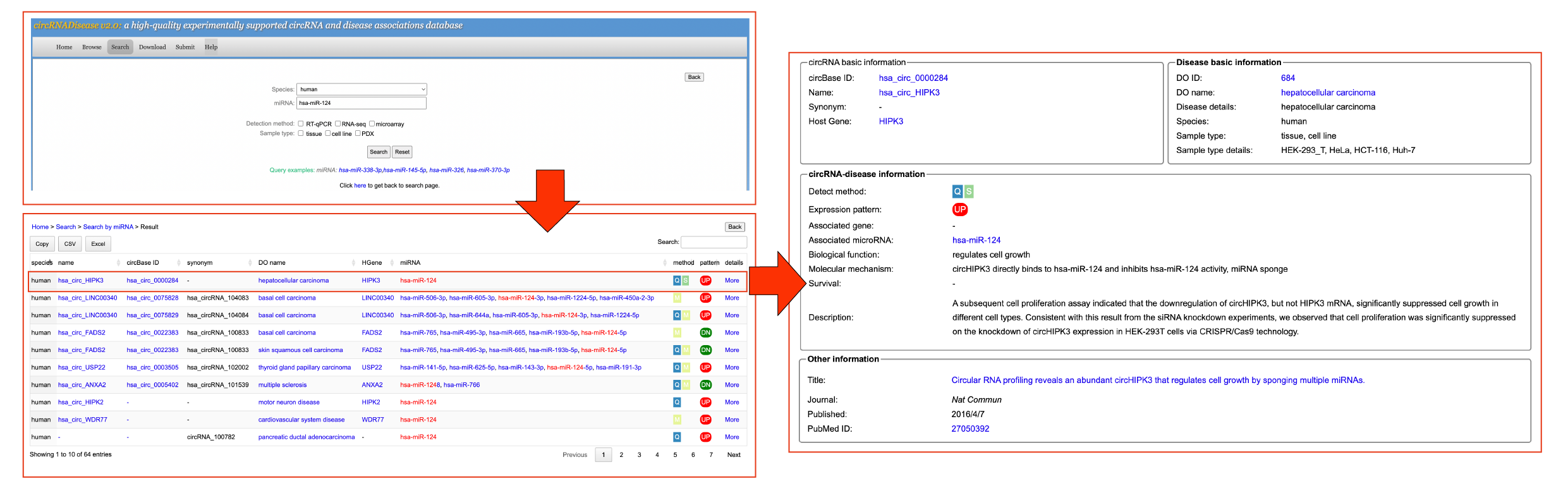
[4.4] Search by miRNAUsers can click the 'Search by miRNA' button and input a miRNA to search the circRNA-disease information.
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
[5] Circ2MutSomatic mutation within non-coding RNAs have also been proven to play critical roles in diseases. Recent studies indicated that mutations in circRNA loci affects circRNAs' biosynthesis or expression. we have collected cancer mutations from the TCGA projects across a broad spectrum of 30 cancer types, encompassing a total of 10,141 samples. Genomic coordinates of those genetic alterations were converted from genome assembly GRCh37/hg19. Finally, we identified 7,417,841 relationships between mutations and circRNAs among 30 TCGA cancer types.
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
[6] DownloadTo download data in the database, please select the menu "Download". The circRNADisease provides two formats of downloadable file in TEXT and Excel formats, respectively.
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
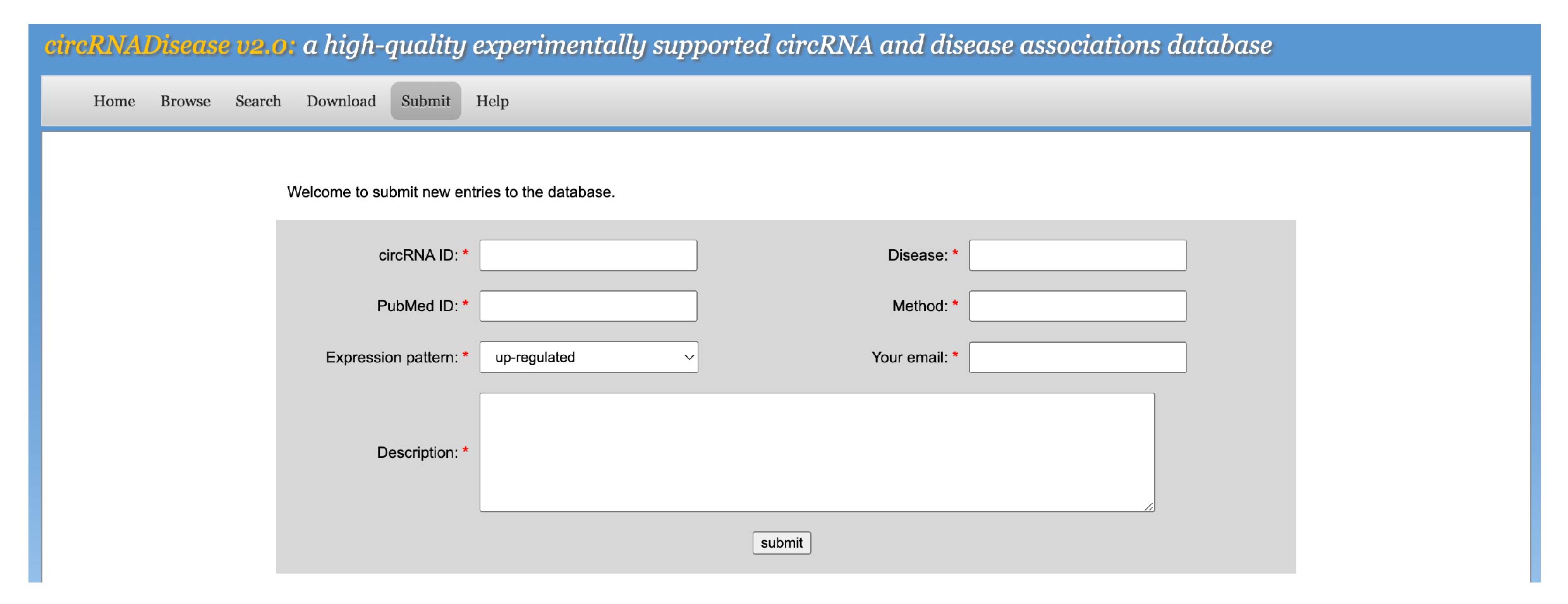
[7] SubmitTo submit data in the database, please select the menu "Submit". We will also encourage research scientists to submit the experimentally supported circRNA-disease association. The users need to input their data into corresponding blanks and then submit the query. The submitted information in the future will be manually reviewed and then integrated into this database.
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
[7] Question and AnswerIf you have any questions, suggestions and comments, please feel free to contact Dr. Zheng Zhao at zhaozheng0503@ccmu.edu.cn |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||